
Documentation
How to run MCLand in your computer?
This page explain how you can set up and run MCLand.
The Python source code for MCLand version 1 for Windows. To run the graphical user interface for MCLand please install Anaconda 3 version 4.3.1 from https://repo.continuum.io/archive/ .
Then use Anaconda Prompt to install python-libsbml package with conda run: conda install -c sbmlteam python-libsbml
Once you have set up the Python 3.6 interpreter, e.g. add the system variable to path C:\ProgramData\Anaconda3, C:\ProgramData\Anaconda3\Scripts, C:\ProgramData\Anaconda3\Library\bin,
you are ready to use Anaconda 3 Spyder (a Python IDE) to open the MCLand_ver1.py file and run the program to start the Graphical User Interface.
1. Introduction
The purpose of this user manual is to guide user on how to use MCLand for plotting and visualizing Waddington's epigenetic landscape for gene regulatory network (GRN). MCLand is a Python software implementing the Monte Carlo method for estimating the probability distribution P(x) and quasi-potential U = - ln P(x) as proposed by Zhang et al. [1]. MCLand enable users to plot and visualize Waddington's epigenetic landscape with ease by using the user-friendly graphical user interface (GUI).
2. Download and Running MCLand
For downloading MCLand please go to the URL: https://github.com/MCLand-NTU/MCLand
2.1 Running MCLand in Windows, Linux or Mac
The Python source code for MCLand Version 1 can be run on Windows, Linux or Mac. Unzip the downloaded MCLand file. For Windows user, the steps for running MCLand are given here. However, for Linux or Mac users the steps should be similar. To run the graphical user interface for MCLand please install Anaconda 3 version 4.3.1 from https://repo.continuum.io/archive/ and then use Anaconda Prompt to install python-libsbml with conda run: conda install -c sbmlteam python-libsbml
Then you have to set up the Python 3.6 interpreter, e.g. add the system variable to path C:\ProgramData\Anaconda3, C:\ProgramData\Anaconda3\Scripts, C:\ProgramData\Anaconda3\Library\bin. After completed the setting, you are ready to use Anaconda 3 Spyder (a Python IDE or Wing 101) to open the MCLand_ver1.py file and run the program to start the Graphical User Interface.
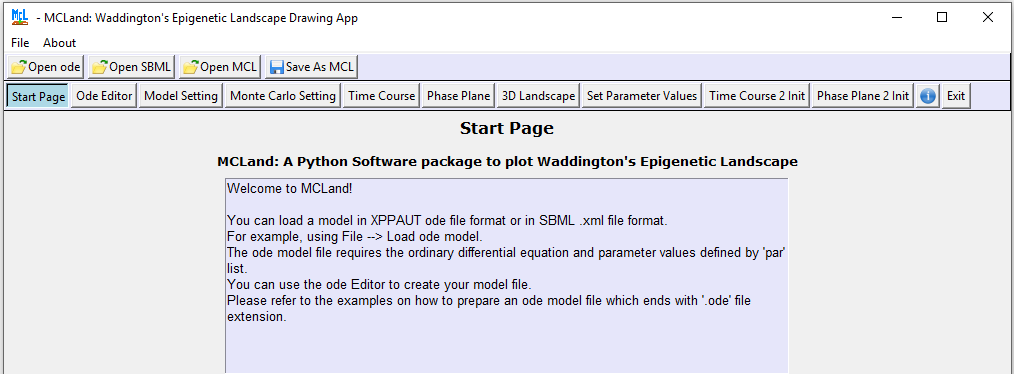
Then the MCLand GUI will be opened as in Figure 1 below.
Reference
1. Zhang, X., Chong, K. H., Zhu, L. & Zheng, J. A monte carlo method for in silico modeling and visualization of waddington's epigeneticlandscape with intermediate details. Biosystems 198, 104275 (2020).